Find optimal preprocessing for your data
One of the most helpful functions in scloop is the ability to sweep over a large amount of embedding parameters at once and compare the results.
We currently offer the following sweep parameters when running the scloop embed command:
CLI argument |
Description |
|---|---|
–preprocessing_sweep |
Test each specified embedding method across all conventional preprocessing techniques and some common combinations of them |
–distance_sweep |
Test each specified embedding method using a range of different distance cutoffs |
–resolution_sweep |
Bin your data to various lower resolutions and run all other settings as provided for each resolution. |
Preprocessing Sweep: Mouse Oocyte/Zygote Dataset
The preprocessing sweep allows you to test each specified embedding method across all conventional preprocessing techniques and some common combinations of them. This is useful for determining which preprocessing method is best for your data, or if an embedding method is biased to a certain representation of the data.
In this example, we demonstrate a preprocessing sweep across both the 1-dimensional and 2-dimensional PCA baseline methods. We can use the following command:
scloop embed --dset oocyte_zygote_mm10 \
--scool data/scools/oocyte_zygote_mm10_1M.scool \
--reference data/oocyte_zygote_ref \
--embedding_algs 1d_pca 2d_pca \
--n_runs 3 \ # need multiple runs to get a good estimate of the variance
--preprocessing_sweep
Running the above command will produce the following clustering accuracy results:
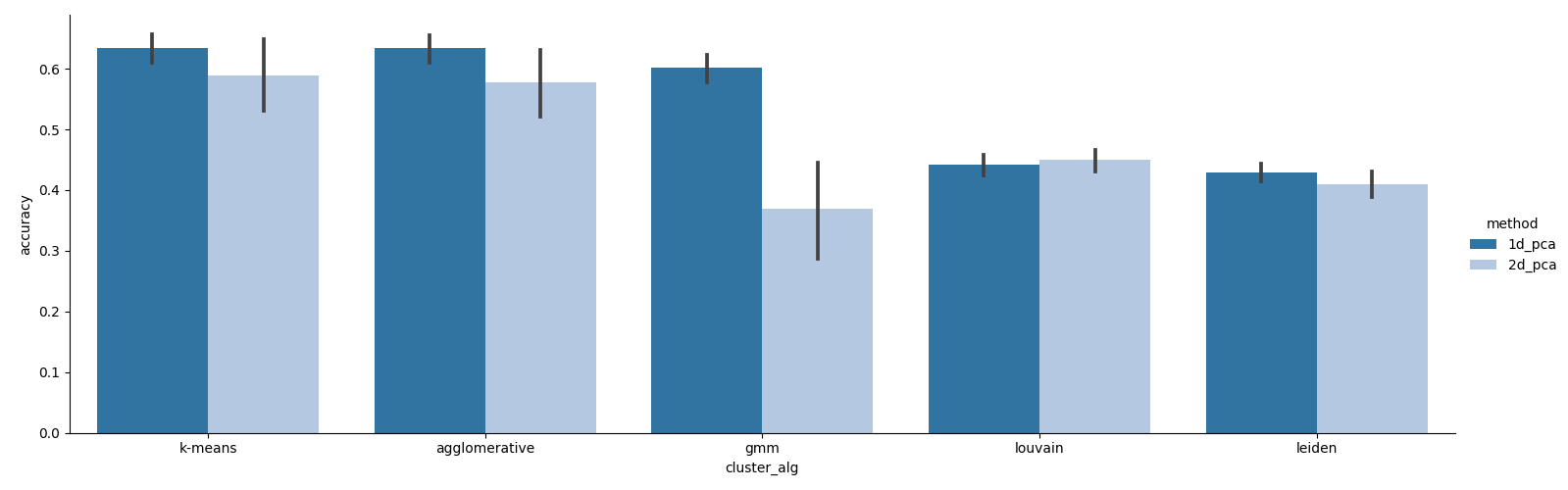
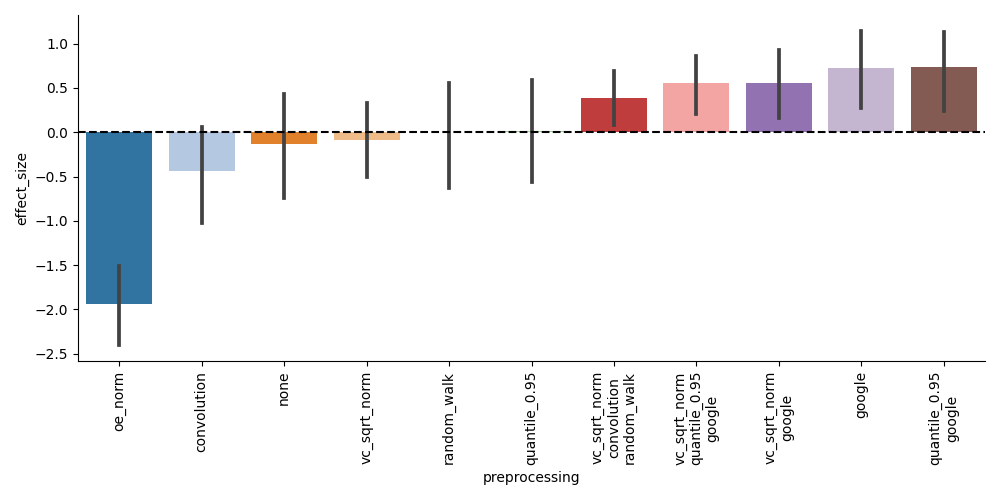
We can see that in general, 2-dimensional representations are not necessary for achieving good clustering accuracy. Indeed, it seems that any 2-dimensional information necessary for embedding these cells can be captured by proper preprocessing. If we look at the effect size of the preprocessing methods, we can see that all of the best-performing preprocessing steps are those that involve coverage correction and graph imputation:
Preprocessing Sweep: Mouse Brain Dip-C Dataset
scloop embed --dset mouse_brain_dipc \
--scool data/scools/mouse_brain_dipc_500kb.scool \
--reference data/mouse_brain_dipc_ref \
--embedding_algs 1d_pca 2d_pca \
--n_runs 3 \ # need multiple runs to get a good estimate of the variance
--preprocessing_sweep
Running the above command, we observe similar results, where 2-dimensional representations are not necessary for achieving good clustering accuracy:
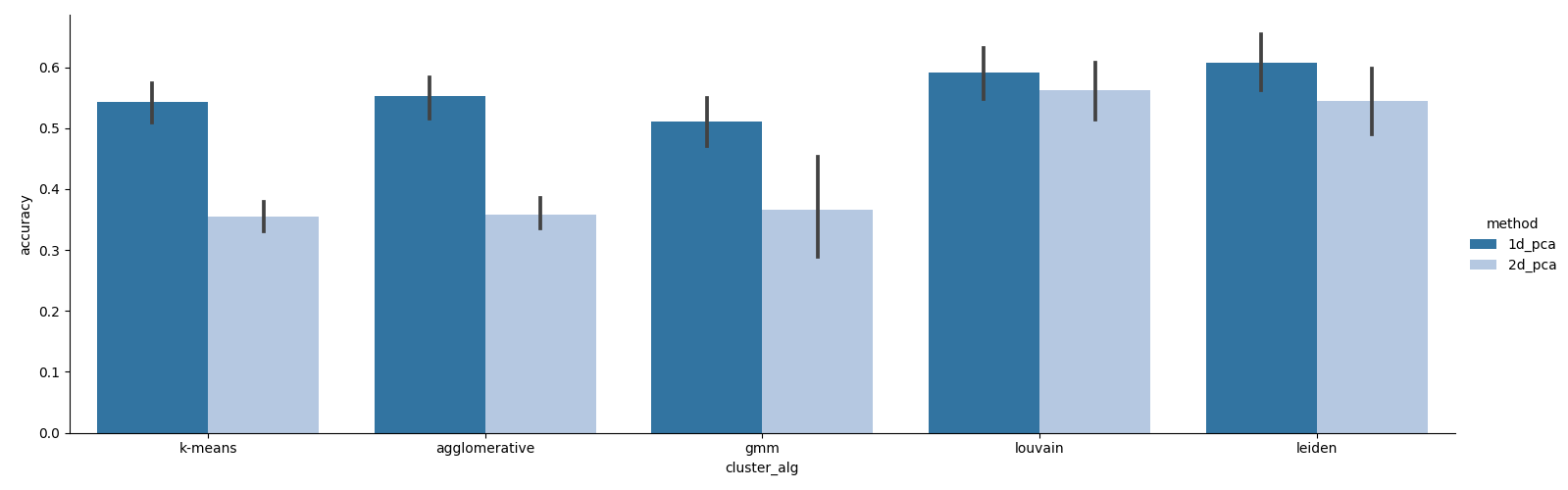
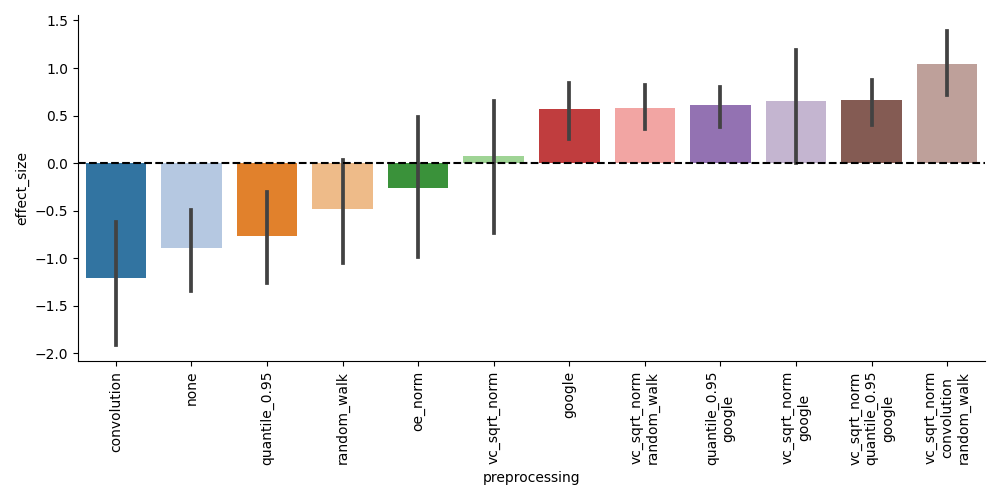
However, this time we see that the best-performing preprocessing steps are only those that involve graph imputation, and not necessarily coverage correction: