Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
About my research
This is a page not in th emain menu
Published:
This post will show up by default. To disable scheduling of future posts, edit config.yml and set future: false.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Training our own re-implementation of the RepNet model, we can achieve superhuman performance in scoring competitive speed jump rope events and even run our model in real-time on an iPhone. See demo here: NextJump and download our training app here: NextJump iOS App 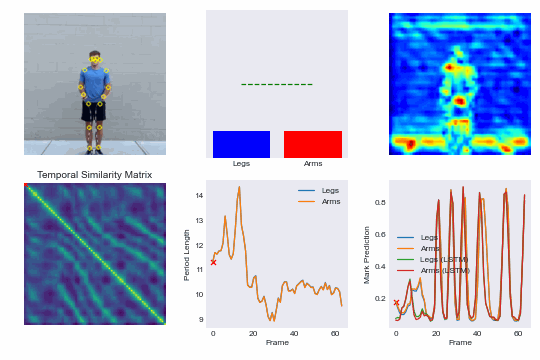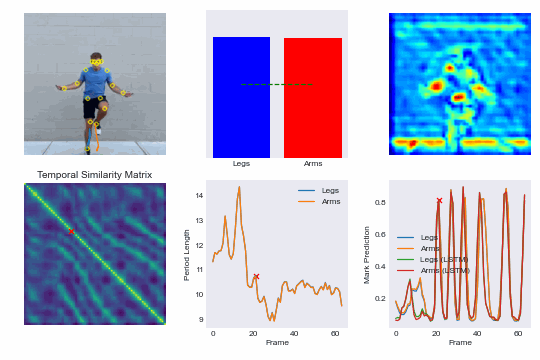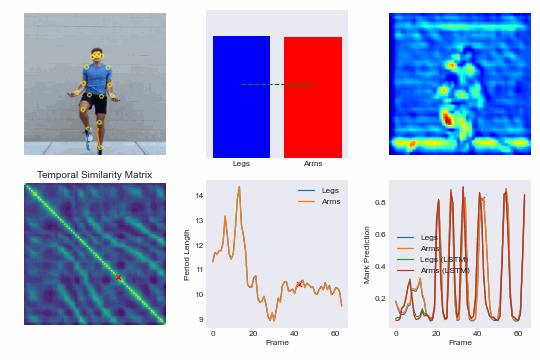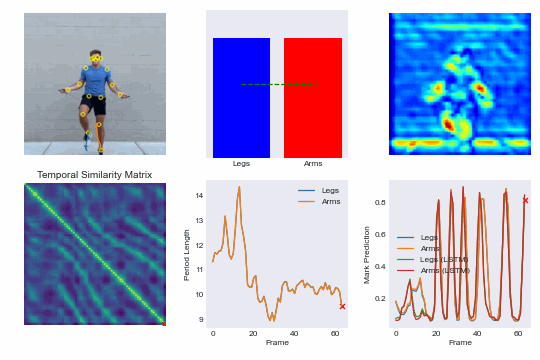
Leading the software subteam for Case Western’s robotics team, CWRUbotix, in our entry into NASA’s annual robotic mining competition.
While jump rope is one of the most physically accessible sports in the world, there exist very few resources for learning advanced skills, especially those needed to compete at an international level. I developed a simple video dictionary of hundreds of skills from basics to impossible which has reached tens of thousands of users across over 60 different countries. 
A Python package currently in development for combining, benchmarking, and extending methods of embedding, clustering, and visualizing single-cell Hi-C data. 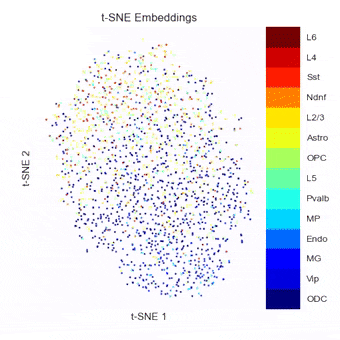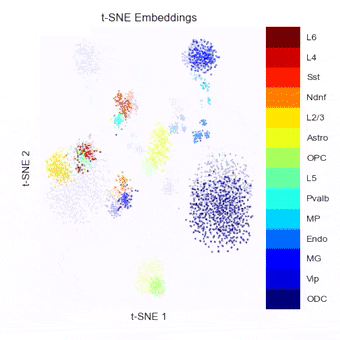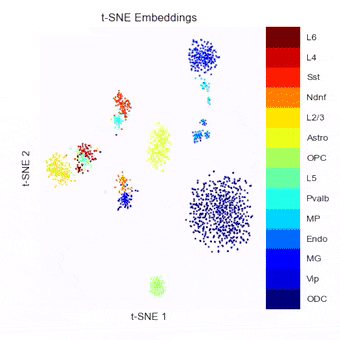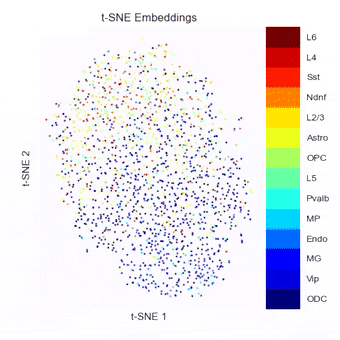
By training a custom RepNet on jump rope videos we can accurately count competitive events. We extend this further by getting our model to run in realtime on iOS devices. This leads to the NextJump app for training jump rope. Download the app here: NextJump App
Published in Nature Genetics, 2022
Mapping chromatin loops from noisy Hi-C heatmaps remains a major challenge. Here we present DeepLoop, which performs rigorous bias correction followed by deep-learning-based signal enhancement for robust chromatin interaction mapping from low-depth Hi-C data. DeepLoop enables loop-resolution, single-cell Hi-C analysis. It also achieves a cross-platform convergence between different Hi-C protocols and micrococcal nuclease (micro-C). DeepLoop allowed us to map the genetic and epigenetic determinants of allele-specific chromatin interactions in the human genome. We nominate new loci with allele-specific interactions governed by imprinting or allelic DNA methylation. We also discovered that, in the inactivated X chromosome (Xi), local loops at the DXZ4 ‘megadomain’ boundary escape X-inactivation but the FIRRE ‘superloop’ locus does not. Importantly, DeepLoop can pinpoint heterozygous single-nucleotide polymorphisms and large structure variants that cause allelic chromatin loops, many of which rewire enhancers with transcription consequences. Taken together, DeepLoop expands the use of Hi-C to provide loop-resolution insights into the genetics of the three-dimensional genome. 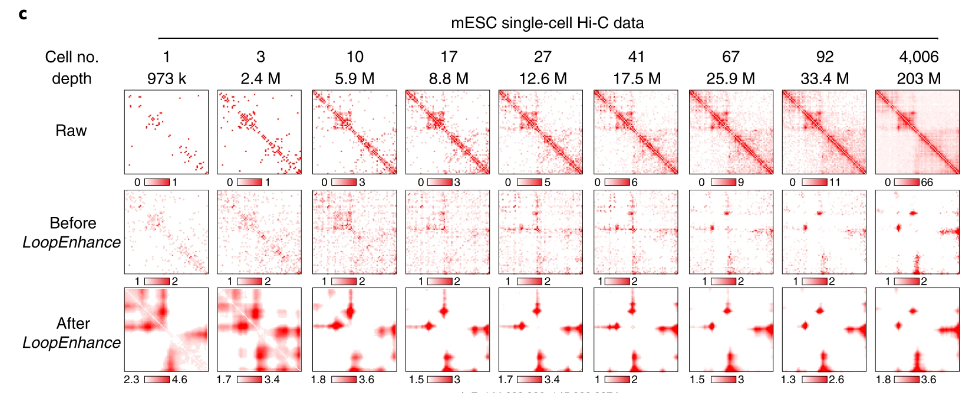
Recommended citation: Zhang, S., Plummer, D., Lu, L. et al. DeepLoop robustly maps chromatin interactions from sparse allele-resolved or single-cell Hi-C data at kilobase resolution. Nat Genet 54, 1013–1025 (2022). https://doi.org/10.1038/s41588-022-01116-w https://www.nature.com/articles/s41588-022-01116-w
Published in Nature Communications, 2023
Broad heterogeneity in pancreatic β-cell function and morphology has been widely reported. However, determining which components of this cellular heterogeneity serve a diabetes-relevant function remains challenging. Here, we integrate single-cell transcriptome, single-nuclei chromatin accessibility, and cell-type specific 3D genome profiles from human islets and identify Type II Diabetes (T2D)-associated β-cell heterogeneity at both transcriptomic and epigenomic levels. We develop a computational method to explicitly dissect the intra-donor and inter-donor heterogeneity between single β-cells, which reflect distinct mechanisms of T2D pathogenesis. Integrative transcriptomic and epigenomic analysis identifies HNF1A as a principal driver of intra-donor heterogeneity between β-cells from the same donors; HNF1A expression is also reduced in β-cells from T2D donors. Interestingly, HNF1A activity in single β-cells is significantly associated with lower Na+ currents and we nominate a HNF1A target, FXYD2, as the primary mitigator. Our study demonstrates the value of investigating disease-associated single-cell heterogeneity and provides new insights into the pathogenesis of T2D.
Recommended citation: Weng, C., Gu, A., Zhang, S. et al. Single cell multiomic analysis reveals diabetes-associated β-cell heterogeneity driven by HNF1A. Nat Commun 14, 5400 (2023). https://doi.org/10.1038/s41467-023-41228-3 https://www.nature.com/articles/s41467-023-41228-3
Graduate course, Case Western Reserve University, Electrical Engineering and Computer Science Department, 2019
Held office hours to answer student questions and graded assignments on basic concepts in Bioinformatics such as:
Graduate course, Case Western Reserve University, Computer and Data Science Department, 2020
Graduate course, Case Western Reserve University, Computer and Data Science Department, 2020
Undergraduate course, Case Western Reserve University, Computer and Data Science Department, 2021
Held office hours to answer student questions and graded assignments on basic concepts in AI such as: