Single cell multiomic analysis reveals diabetes-associated β-cell heterogeneity driven by HNF1A
Published in Nature Communications, 2023
Recommended citation: Weng, C., Gu, A., Zhang, S. et al. Single cell multiomic analysis reveals diabetes-associated β-cell heterogeneity driven by HNF1A. Nat Commun 14, 5400 (2023). https://doi.org/10.1038/s41467-023-41228-3 https://www.nature.com/articles/s41467-023-41228-3
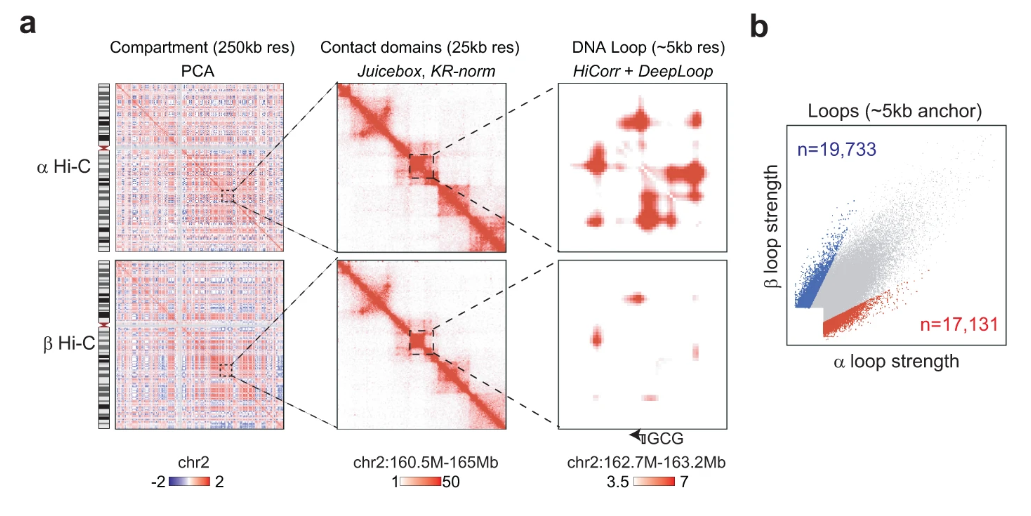
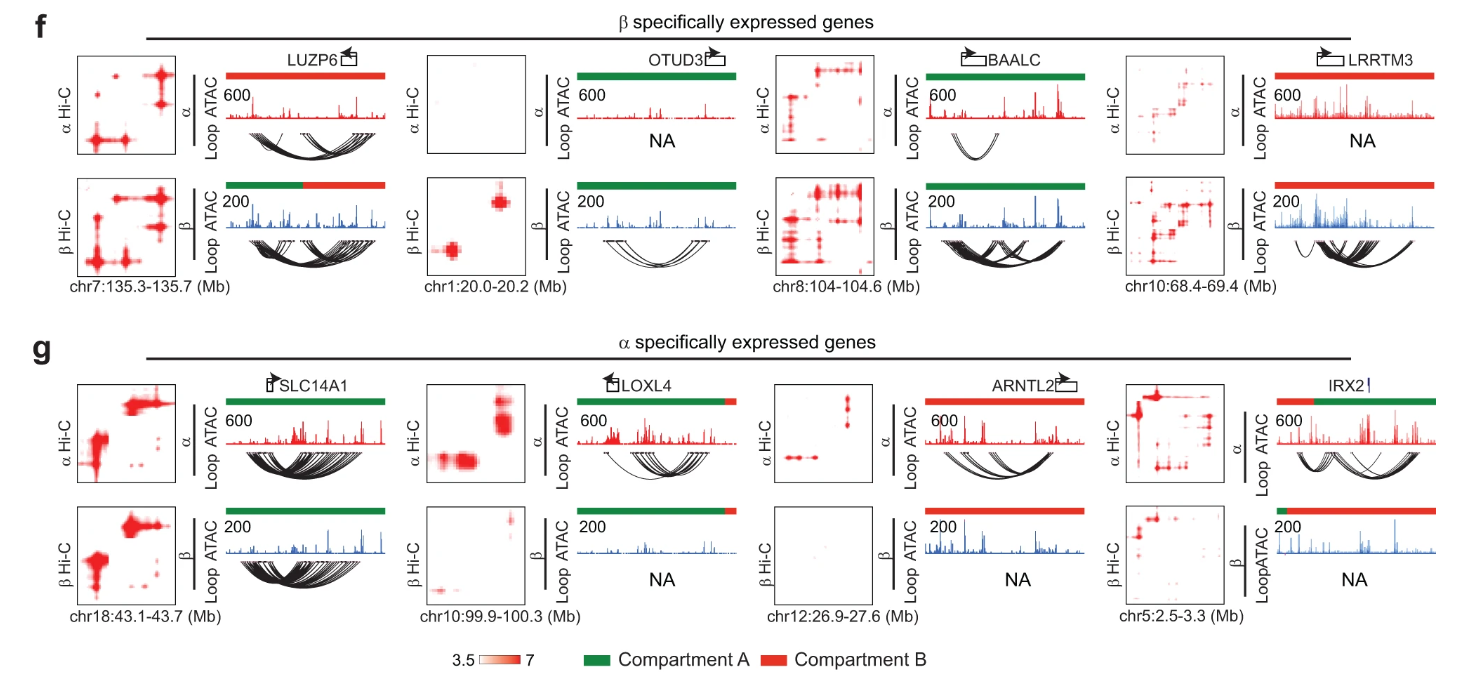
In this work, our lab investigates many aspects of pancreatic islet heterogeneity across different single-cell modalities (transcription and chromatin accessibility profiles) between celltypes and within β-cells specifically. My main contributions involve utilizing our DeepLoop pipeline for analyzing bulk Hi-C data to identify β-specific and α-specific loops from high resolution eHi-C data.
While compartment and TAD structures are highly similar between these two celltypes, we find subtle loop-level differences which can be further confirmed through celltype specific analysis of the chromatin accessibility peaks identified by multiomic analysis.
These results demonstrate the practical feasibility of the DeepLoop pipeline in analyzing highly specific chromatin loops.