Dylan’s Research Page
About Me
I am Dylan and I perform research at the intersection of machine learning and genomics. I received my PhD from Case Western Reserve University advised by Jing Li and Fulai Jin. Beyond research, I am also a world champion jump roper building computer vision applications for sport analysis and competition scoring.
About my Research
I believe that the main frontier of the future of machine learning will be in the natural sciences, and I see biology—specifically genomics—as the most important, interesting, and currently accessible of those fields. Data-driven genomics and machine learning have formed a symbiotic relationship and both fields will continue to advance through interdisciplinary research.
Thanks to incredible developments in next-generation sequencing technology, experiments exist to profile complex epigenetic states such as the 3-dimensional structure of the genome. Single-cell protocols are able to profile these epigenetic features in individual cells. The data produced by these experiments is high dimensional, noisy, and differs from typical transcriptional data like RNA-seq because the features are more complex and indirectly resposible for cell states than just gene expression, thus celltype heterogeneity of the epigenome and its link to transcription is not well understood. My current research mainly involves determining the best approach for embedding this data and how low dimensional representations can be used to discover novel biology, focusing on the relationship between the epigenome and the transcriptome via 3D genome structure.
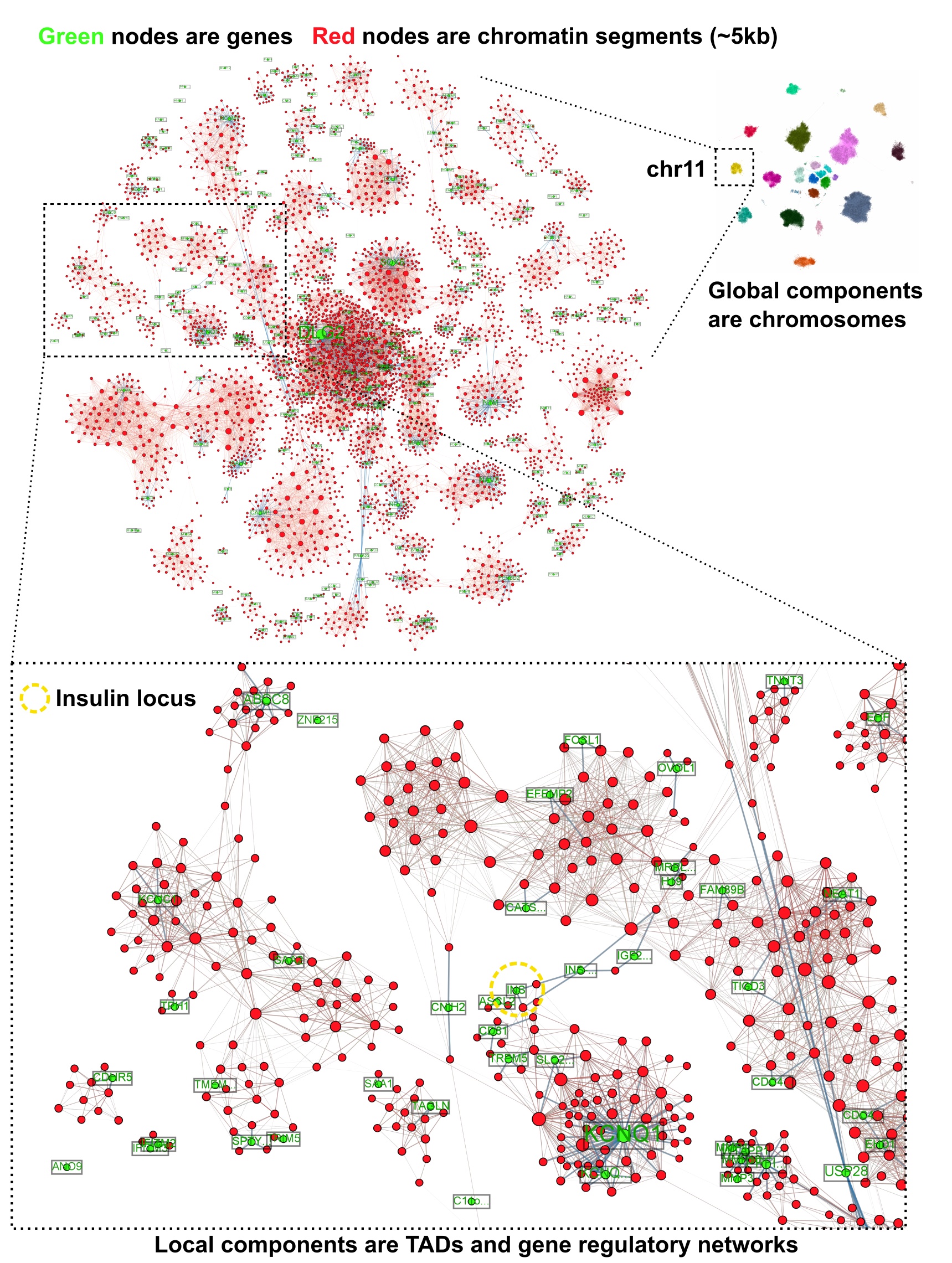
Currently I work with a lot of Hi-C and single-cell datasets, and these often require developing novel machine learning techniques for taking advantage of the unique structure of the data and the biological priors we might already have. My goals are to develop computational tools that allow biologists to disentangle the epigenome and yield further insights into development and disease genetics. I envision our field developing and iterating upon these various tools (single-cell representation and integration methods, DNA/RNA/protein language models, spatial single-cell analysis techniques, etc) in tandem with new protocols for generating data, eventually converging on comprehensive models of the dynamic genome which can be queried, perturbed, and simulated in larger biological systems.
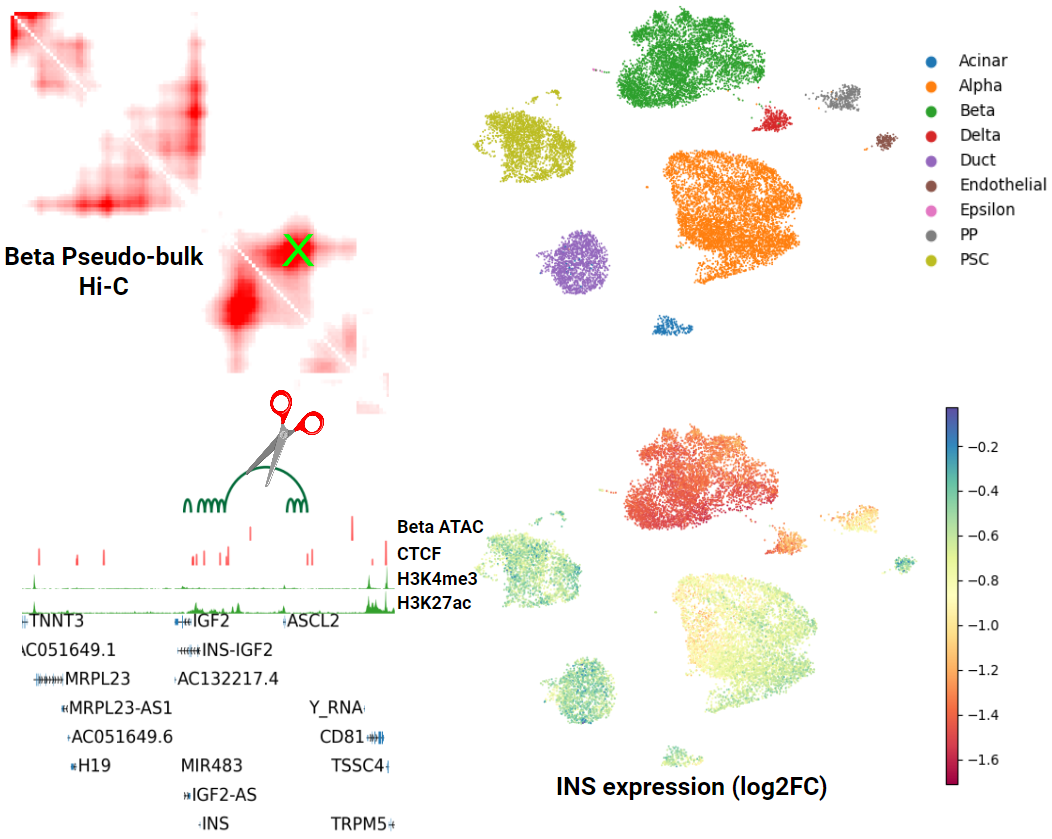
One of my main focuses right now is building generative models of the epigenome and its relationship to transcription. Below is an example of one of my models measuring the effect of an individual chromatin loop perturbation on gene expression via a joint embedding of scRNA-seq and scHi-C data.
This is done through graph representation learning on a massive genome-wide guidance graph. Generative modeling with biologically-informed priors such as these is how I envision the future of deep learning in genomics.